Journal of Creation 18(1):11–12, April 2004
Browse our latest digital issue Subscribe
Bone building: perfect protein
Bones are an amazing example of design, present in all vertebrates. They have a huge advantage over man-made girders, in that they are constantly rebuilding and redesigning themselves to cope with changing stress directions.1
This involves a fine balance of the activity of bone-depositing cells (osteoblasts) and bone resorbing cells (osteoclasts). It’s been recently shown that thyroid-stimulating hormone (TSH), best known for what its name says—stimulating the production of hormones in the thyroid gland—has an important role. It oversees both types of cells—without it, bones have osteoporosis in some parts (too little bone, so very weak), and are too dense in other patches.2
Osteocalcin and hydroxyapatite
The strength of bones mainly comes from the hexagonal mineral hydroxyapatite (HA, formula Ca₁₀(PO₄)₆(OH)₂).3 But this must be built up in the right patterns. In vertebrate bones, this is built up with a special protein called osteocalcin (OC). It is a small protein, 49 amino acids long (5.8 kDa), and is ‘highly conserved’, meaning that its sequence is almost identical among vertebrates. Human OC has the sequence Tyr Leu Tyr Gln Trp Leu Gly Ala Pro Val Pro Tyr Pro Asp Pro Leu Gla Pro Arg Arg Gla Val Cys Gla Leu Asn Pro Asp Cys Asp Glu Leu Ala Asp His Ile Gly Phe Gln Glu Ala Tyr Arg Arg Phe Tyr Gly Pro Val.4
Like all proteins, the instructions for OC are in the DNA,5 but there is more to its manufacture than simply decoding / translating the code and synthesizing the OC on a ribosome. Firstly, the transcription (DNA→mRNA) is regulated by 1,25-dihydroxy-Vitamin D3, one reason that Vitamin D is so important for healthy bones. It is then first decoded (translated) as a preproosteocalcin, which is 98 amino acids long. This comprises three parts: a 23-residue signal protein that is cleaved during translation,5 a 26-residue target propeptide, and the 49-residue mature protein.6
Even this does not complete the process; this requires another vitamin—K. Vitamin K1 or phylloquinone, best known for its vital role in the blood clotting cascade, is an essential co-factor in γ-carboxylation. That is, the specific glutamyl residues (Glu, from the amino acid glutamic acid) at positions 17, 21 and 24 have a second carboxyl group (–COOH) added to form γ-carboxyglutamyl residues (Gla). This changes the structure and stabilizes the α-helical portion of the protein.6
Even now, the OC protein is fairly shapeless. But when OC meets calcium ions, it folds to a special structure.8 The two carboxyl groups on the γ-carboxyglutamyl residues chelate1 to the Ca2+ ions, as shown by Fourier-Transform Infrared spectroscopy (FTIR).9 There was no spectral change when Ca2+ was added to decarboxylated OC (i.e. as it would be before converted by Vitamin K), showing that there is no binding without the carboxylation.9 Amazingly (for uniformitarians), enough osteocalcin to produce an immune reaction was found in bones of an Iguanodon ‘dated’ to 120 Ma,10 yet proteins could not last for millions of years. And the fact that it’s a bone protein shows it can’t be contamination from outside.
Osteocalcin’s crystal structure
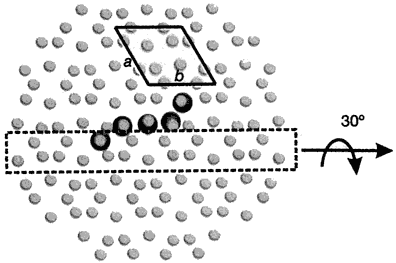
Now, pig OC’s crystal structure has been discovered, using a type of X-ray diffraction called the iterative single anomalous scattering method. This provides new insights into how finely designed it must be to work.7 The active site has a negatively charged region that binds the positively charged Ca2+ ions. Five Ca2+ ions are coordinated by three special Gla residues and an Asp at position 30. But not in just any old way—five calcium ions are bound in the same arrangement as in the exposed face of a HA crystal, parallel to the c axis. So the OC can dock on the HA and add the calcium, and thus grow the crystal, making the bone grow in the area needed.
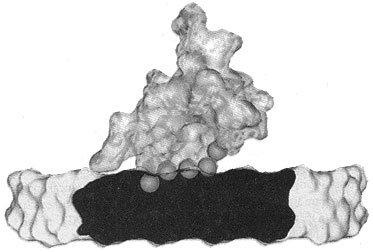
To do this, OC’s building blocks, the amino acids, must be in a very precise sequence. For example, there is a tightly packed core involving the hydrophobic residues Leu 16, Leu 32, Phe 38, Ala 41, Tyr 42, Phe 45 and Tyr 46. There is also hydrogen bonding to stabilize the connection between different α-helices, Asn 26 in the helix α1–α2 linker and Tyr 46 in α3. The helices α1 and α2 form a V-shaped arrangement stabilized by a disulphide bridge between Cys 23 and Cys 29.
Thus the very precise sequence of OC, as well as the metabolism to form the essential γ-carboxyglutamyl residues, seems to be yet another example of irreducible complexity, a hallmark of design.11 This means this is yet another component that must be exactly right for the alleged transition from invertebrate to vertebrate. So it is not surprising that evolutionists have no fossil evidence for how the transition occurred—this protein alone shows it could never have happened.12
Summary
- Bones are dynamic supports, constantly rebuilding to cope with changing stresses.
- Bone shape is a delicate balance of bone deposition and resorption.
- Bone strength is largely from the mineral hydroxyapatite (HA).
- One vital component for bone growth is the small, highly conserved protein osteocalcin (OC).
- Large fragments have been found in dinosaur bones ‘dated’ over 100 million years old, although measured rates of breakdown mean that nothing should have survived that long.
- Vitamin K is essential to modify three amino acid residues of OC, otherwise it can’t bind calcium at all.
- Recent discovery of OC’s crystal structure shows that it binds calcium in exactly the right geometry to add to a certain crystal face of HA.
- Thus bone construction is irreducibly complex.
- Which explains why there is not only no fossil intermediate between invertebrate and vertebrate, but why it could not exist (and mainly because God didn’t create one!).
References
- Wieland, C., Bridges and bones, girders and groans, Creation 12(2):20–24, 1990; creation.com/bones. Return to text
- Tromans, A., Foreman in the bone factory, Nature 425(6961):909, 30 October 2003. Return to text
- Space group P63 /m (C6h2), unit cell dimensions a = b = 9.432 Å, c = 6.881 Å; Kay, M.I., Young, R.A. and Posner, A.S., The crystal structure of hydroxyapatite, Nature 204:1050–1052, 1964. Return to text
- American Peptide Company, Inc., Osteocalcin (1-49), Human, www.americanpeptide.com, 2002. Return to text
- Sarfati, J., DNA: marvelous messages or mostly mess? Creation 25(2):26–31, 2003; creation.com/message. Return to text
- Lee, A.J., Hodges, S. and Eastell, R., Measurement of osteocalcin, Annual of Clinical Biochemistry 37:432–436, 2000; www.leeds.ac.uk/acb/annals/annals_pdf/July00/432.PDF. Return to text
- Hoang, Q.Q. et al., Bone recognition mechanism of porcine osteocalcin from crystal structure, Nature 425(6961):977–980, 30 October 2003. Return to text
- To chelate means one ligand binds in two or more places to one metal ion, like a claw (Greek χηλή chēlē). Return to text
- Mizuguchi, M., Fujisawa, R., Nara, M., Nitta, K. and Kawano, K., Fourier-Transform Infrared Spectroscopic Study of Ca2+-Binding to Osteocalcin, Calcified Tissue International 69(6):337–342, December 2001. Return to text
- Embery G., Milner A.C., Waddington R.J., Hall R.C., Langley M.S., Milan A.M., Identification of proteinaceous material in the bone of the dinosaur Iguanodon, Connect Tissue Res. 44 Suppl 1:41–6, 2003; www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12952172&dopt=Abstract. The abstract says: ‘an early eluting fraction was immunoreactive with an antibody against osteocalcin.’ Return to text
- A term made famous by Behe, M.J., Darwin’s Black Box: The Biochemical Challenge to Evolution, The Free Press, New York, 1996. Return to text
- See Gish, D.T., Evolution: The fossils STILL say NO!, pp. 74–76, Institute for Creation Research, El Cajon, CA, USA, 1995. Return to text


Readers’ comments
Comments are automatically closed 14 days after publication.